Background
Cancer cells have prodigious anabolic and energetic requirements and use distinct metabolic pathways
for growth and survival. In addition to glucose, glutamine plays an important role in providing both
carbon and nitrogen necessary for anabolic metabolism. As glucose is metabolized through glycolytic
pathways to lactic acid, glutamine can fuel the TCA cycle, generating metabolic intermediates to serve as
building blocks for lipids, proteins, and nucleic acids that are crucial to anabolic growth and
proliferation. Notably, these same metabolic programs have also been attributed to facilitating the
tremendous growth associated with T cell activation and proliferation. An important consequence of the
specialized metabolism of cancer cells is the creation of a hypoxic, acidic, nutrient-depleted tumor
microenvironment (TME) that is hostile to antitumor immune responses. In other words, the specialized
metabolic programming of cancer cells not only promotes tumor growth but can also block the
generation of an effective anti-tumor immune response. Therefore, in clinical, the metabolic
characteristics of tumors present considerable hurdles to immune cell function and cancer
immunotherapy.
We hypothesized that blocking glutamine metabolism would not only inhibit tumor growth but also relieve a master immunometabolic checkpoint and enable restoration of antitumor immunity. However, the therapeutic response of tumors in vivo to targeted blockade of glutaminase is generally limited. Instead of inhibiting a single enzyme, we chose to comprehensively inhibit glutamine metabolism by using a pro-drug of a glutamine antagonist. In addition to glutaminase, this compound inhibits a broad range of glutamine-requiring enzymes.
How we answered this Question
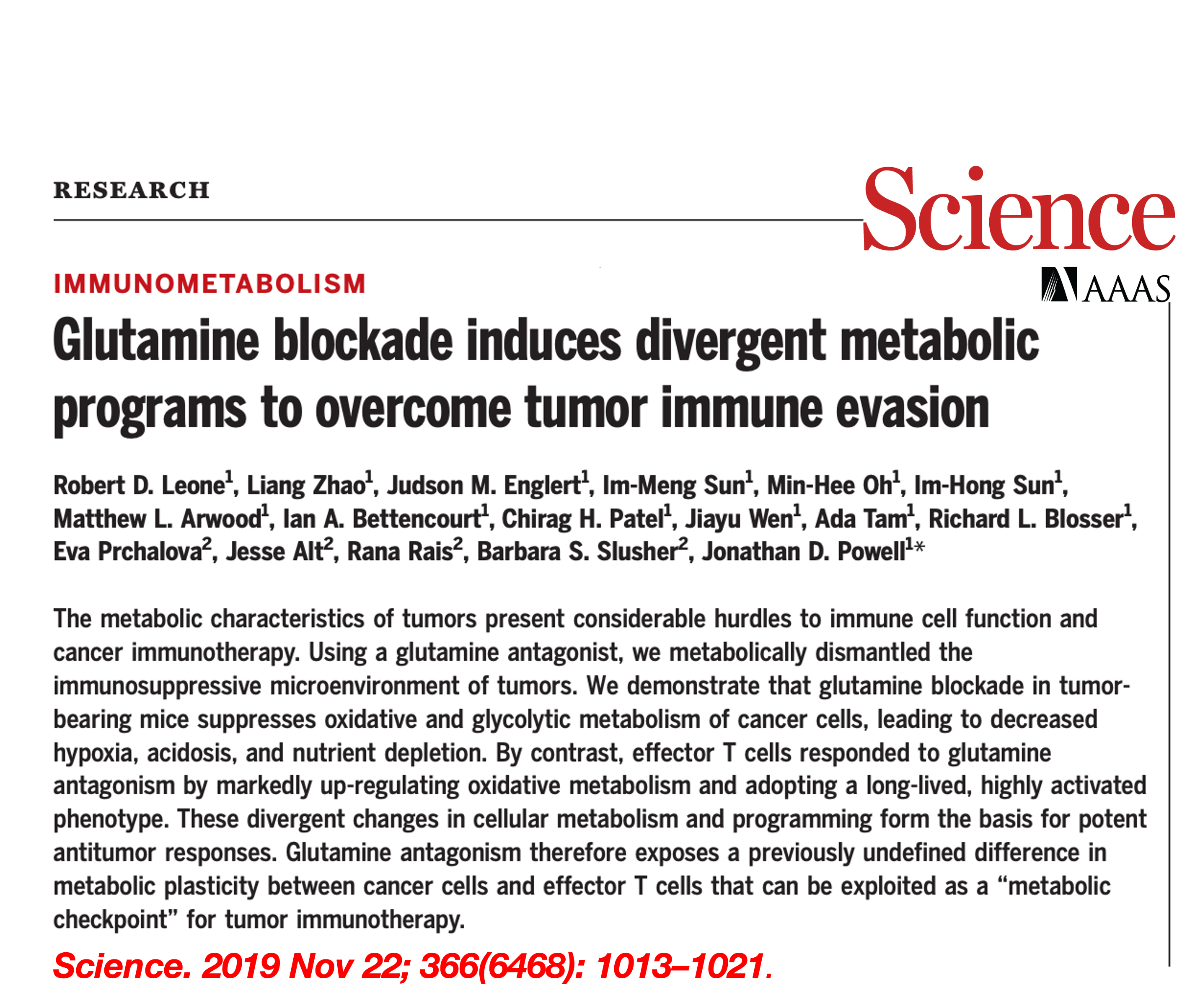
To examine the ability of this new drug to inhibit tumor growth, we performed a series of experiments
using different syngeneic tumor models. Specifically, we used the MC38 in vivo model to examine the
specific effects on tumor metabolism. Using 13 C-glucose tracing approach, we observed broad
suppression of metabolic activity after drug treatment (Fig. 1).
While genomics, transcriptomics, and proteomics provide valuable information, they can be limited in providing a definitive representation of the clinical phenotype. In contrast, metabolomics provides a snapshot of an organism’s current metabolic status and also provides actionable information that can help advance research and clinical decision-making. Therefore, the phenotyping power of metabolomics makes it an ideal tool for studying the molecular mechanisms of disease, and metabolomics plays important roles in many fields. In addition, metabolomics links genotype to phenotype. This linkage supports the interpretation of other ‘omics data that, when considered alone, may not translate in the observed phenotypes, resulting in misinterpretation.
—Dr. Liang Zhao
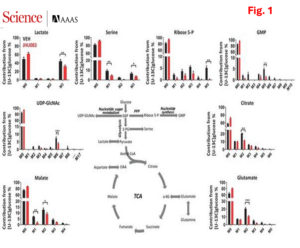
Such findings suggest that blocking glutamine metabolism can critically disrupt the metabolism of the tumor as a whole, with marked effects on the nutrient milieu within the TME. More interestingly, our results indicate that blockade of glutamine metabolism in tumor, does not metabolically disable antitumor immune cells, and appears to robustly enhance their function and effectiveness. Based on this finding, we hypothesized that T cells used an alternative anaplerotic source to maintain TCA activity. To test this, we performed a series of in vitro stable-isotope tracing studies, we found that drug-treated MC38 tumor cells show a profound suppression of glucose-derived carbons contributing to the TCA cycle. In contrast, drug treated T cells show considerable flexibility in glucose metabolism, glucose-derived carbons, both as acetyl-CoA as well as oxaloacetate through PC-mediated anaplerosis (Fig. 2).